Definition 1.4
A 1-
PAM unit is the amount of evolution which will change,
on average,
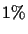
of the amino acids. In mathematical terms, this
is expressed as a matrix
M such that

,
where
fi is the frequency of the
ith
amino acid and
Mij is the probability that an amino acid
i mutates into an amino acid
j
at one PAM distance [
5]. The function
PAM(s1,
s2) is the PAM distance of two sequences
si,
s2 that
maximizes the OPA score.