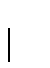 of the amino acids. In mathematical terms, this is
expressed as a matrix M such that
of the amino acids. In mathematical terms, this is
expressed as a matrix M such that
The definition of matrix M describes mutation over a given period of
evolution. In order to procede, we must quantify this change in
a mathematically meaningful way. Dayhoff et. al.[9]
introduced the term PAM (point accepted mutation) unit. A
1-PAM unit is the amount of evolution which will change, on
average, 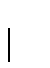 of the amino acids. In mathematical terms, this is
expressed as a matrix M such that
of the amino acids. In mathematical terms, this is
expressed as a matrix M such that
If we have a probability or frequency vector p, the product ![]() gives the probability vector (or the expected frequency of p) after
an evolution equivalent to 1-PAM unit.
gives the probability vector (or the expected frequency of p) after
an evolution equivalent to 1-PAM unit.
Alternatively, if we start with amino acid i (a probability vector
which contains a 1 in position i and 0s in all others),
 (the ith column of M) is the corresponding
probability vector after one unit of random evolution.
(the ith column of M) is the corresponding
probability vector after one unit of random evolution.
After k units of evolution (a k-PAM evolution), a frequency
vector p will be changed into the frequency vector
![]() .
.
Note that PAM distance does not correlate in any immediate way to chronological time. Evolutionary rates may be very different between species and proteins.