![[*]](cross_ref_motif.gif) - Randomization,
Statistics and Visualization contain a complete description of
options and protocols for graphic output.
- Randomization,
Statistics and Visualization contain a complete description of
options and protocols for graphic output.
The function DrawSimPam produces a graph of the similarity score against different PAM distances for a given alignment. This gives the user an intuitive notion of how the similarity changes with changing distances, how precisely defined the maximum is, etc. For example:
> DB := ReadDb('~cbrg/DB/SwissProt'): CreateDayMatrices():
> m := Match(op(Sequence(Entry(8588))), op(Sequence(Entry(8577)))):
> m := LocalAlignBestPam( m, DMS ):
> DrawSimPam(m);
The maximum, 275.4, is located at pam=84
The results are contained in Figure ![[*]](cross_ref_motif.gif) .
.
Like all Darwin functions producing graphic files, the results of a
DrawSimPam are stored by default in the file temp.ps in the
current directory. Chapter ![[*]](cross_ref_motif.gif) - Randomization,
Statistics and Visualization contain a complete description of
options and protocols for graphic output.
- Randomization,
Statistics and Visualization contain a complete description of
options and protocols for graphic output.
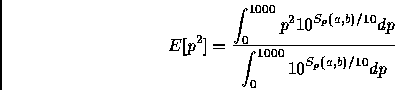 |