


Next: Bibliography
Up: Evaluation Measures of Multiple
Previous: Example
We have defined a new scoring function, called CS measure, for the
evaluation of MSAs that is based on a Markovian model of
evolution. The frequently used SP measure, which calculates the
sum of the scores of all pairwise alignments, ignores the
structure of any associated phylogenetic tree and thus weights
some evolutionary events more than others. This can be avoided by
traversing the tree in a circular order, where all edges are
traversed exactly twice. Any other non-circular tour results in a
longer path, because some edges are traversed more than twice, and
hence some evolutionary events are counted more often, which
corresponds to a lower probability. Therefore the shortest cycle
through the tree, which is a circular tour, yields the highest CS
score. Such a tour can be calculated with any TSP algorithm. The
The TSP application allows us to avoid the calculation of an
evolutionary tree altogether, and only the sequences at the leaves
are needed with their OPA or MPA scores. In addition, the CS
measure yields a direct connection between an MSA and the
associated evolutionary tree, because the formula for the
calculation of the score is exactly the same. The CS measure
based on the OPA scores gives an upper bound on the score of the
optimal MSA and evolutionary tree.
To illustrate the new scoring tool we simulated evolution by
generating random trees according to a Markovian model with the
corresponding MSA. The CS score of the generated alignment was
then compared to the score of alignments calculated by different
methods (MSA, ClustalW, PAS, and MAP). For less than twenty
sequences MSA and PAS gave the best score, whereas for larger
samples PAS and ClustalW calculated the best alignments. A better assessment must come with
actual experimental sequence data though.
With the new CS measure we have now the possibility of improving
current algorithms and finding new algorithms by maximizing the
scoring function. The most simple (and expensive) approach would
be a standard dynamic programming algorithm. An approximation
algorithm is presented in [Korostensky and Gonnet, 1999a] that calculates an MSA
which is within
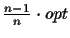 (where opt is the
optimal score). The CS scoring function can also be used for
evolutionary trees, and a tree construction algorithm that is
presented in [Korostensky and Gonnet, 1999b]. The algorithms have been implemented
into the Darwin system [Gonnet, 1994b] an are available via
our cbrg server at:
(where opt is the
optimal score). The CS scoring function can also be used for
evolutionary trees, and a tree construction algorithm that is
presented in [Korostensky and Gonnet, 1999b]. The algorithms have been implemented
into the Darwin system [Gonnet, 1994b] an are available via
our cbrg server at:
http://cbrg.inf.ethz.ch



Next: Bibliography
Up: Evaluation Measures of Multiple
Previous: Example
Chantal Korostensky
1999-07-14