


Next: Tree construction using gaps
Up: Methods
Previous: Improving Alignments With Gap
We did a large simulation where more than 1000 MSAs with 16, 20 and 30 sequences
were randomly generated by simulating evolution. The length of
each sequences was around 200. The score of the generated MSA was
noted. The unaligned sequences were then given to two different
algorithms, ClustalW [11] and ProbModel
[15]. The resulting MSAs were scored and then given to
our new algorithm to improve the alignments. The difference in score of the final alignment and the
original alignment was noted. We also counted how often our algorithm improved the MSAs.
The results are depicted in Table
![[*]](/pub/latex2html/1.1p1/icons.gif/cross_ref_motif.gif) . In all cases, our gap algorithm could improve at
least
. In all cases, our gap algorithm could improve at
least 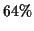 of the MSAs.
of the MSAs.
Figure:
The columns ProbModel and ClustalW show the percentage of
MSAs that have improved. The column before is the percentage
of MSAs that scored at least as well as the real alignment before
the improvement, the column after is the percentage after the
gap algorithm was applied.
 |
Chantal Korostensky
1999-07-14

